Introduction
pearGRN (Pear Integrative Gene Regulatory Network Database) is a comprehensive resource designed to consolidate various omics regulatory networks, facilitating the retrieval of gene regulatory relationships and protein interactions. It provides a platform to predict key genes associated with important breeding-related traits.
The database offers several tools including BLAST, Expression, Jbrowse, Gene Search, Node Search, Network Creation, Network Comparison, TF Regulation, Interact Finder, TraitConnect and ML Predictor, making it an essential resource for researchers in pear genetics and genomics.
PearGRN is hosted by Central of Pearl Engineering Technology Research at Nanjing Agricultural University and aims to support advanced genetic research and breeding projects in pears.
Citations:
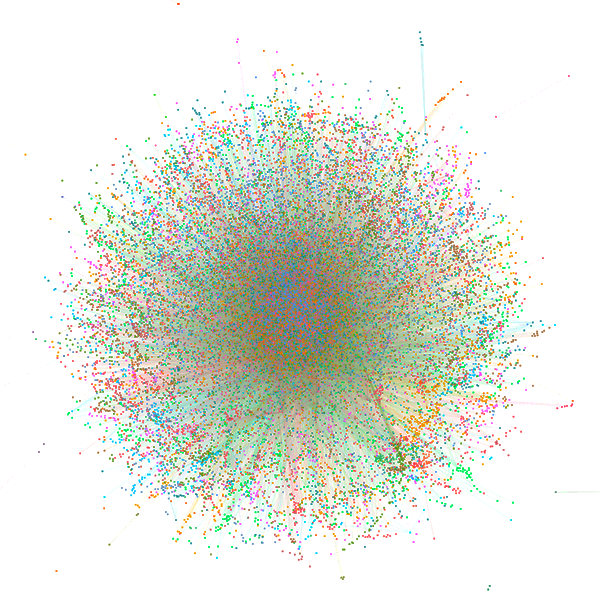
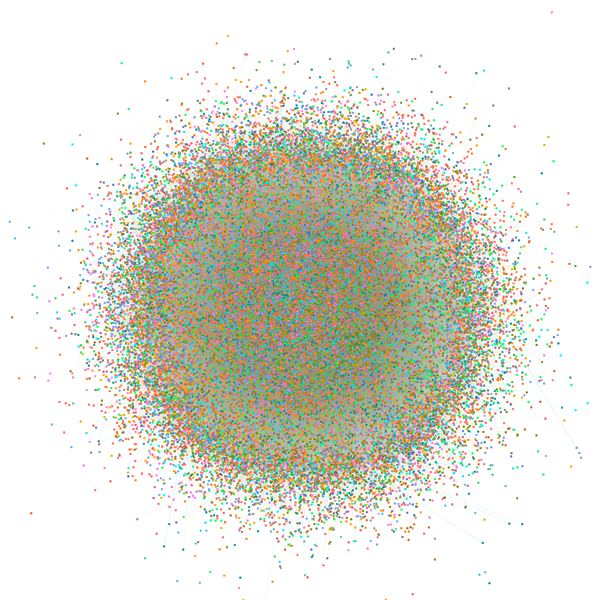
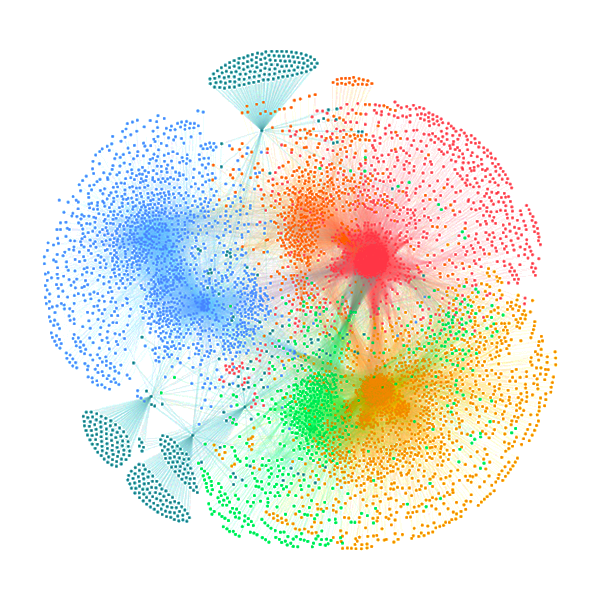
Network Summary Information
| Network | Node number | Edge number | Transitivity | Avg. of distance | Modules (>=5 genes) | Hub gene |
|---|---|---|---|---|---|---|
| Fat CoE Network | 35007 | 2658240 | 0.371 | 2.75 | 678 | 3496 |
| Slim CoE Network (HC) | 29723 | 351635 | 0.283 | 4.09 | 1693 | 2955 |
| Slim CoE Network (LC) | 31929 | 3896398 | 0.302 | 2.67 | 422 | 3192 |
| PWM Network | 39473 | 395509 | 0.004 | 2.61 | 101 | 3799 |
| ATAC-PWM Network | 9717 | 194238 | 0.012 | 2.51 | 22 | 956 |
| Pcorr Network | 2304 | 269130 | 1 | 1 | 21 | 492 |
| cPPI Network | 6790 | 64003 | 0.117 | 3.43 | 269 | 670 |
| 3D Inter Network | 3032 | 5237 | 0.019 | 2.59 | 228 | 261 |
| 3D Intra Network | 8591 | 16743 | 0.045 | 3.75 | 559 | 805 |
| 3D Network | 10964 | 21980 | 0.05 | 4.55 | 737 | 1059 |
| Nocoding Network | 25700 | 1626783 | 0.8 | 5.24 | 929 | 2484 |
| Interactome Network | 8666 | 333115 | 0.944 | 3.31 | 262 | 825 |
| TF-target Regulatory Network | 44574 | 1019312 | 0.015 | 2.56 | 761 | 4425 |
| Integrative Network | 45933 | 2871763 | 0.257 | 2.57 | 926 | 4586 |